Bayesian R2 and LOO-R2
Aki Vehtari, Andrew Gelman, Ben Goodrich, Jonah Gabry
First version 2019-01-02. Last modified 2025-11-12
Update 2025-11-12: loo_R() function definition had a typo which has been fixed. This change has a negligible effect in LOO-R2 results and no effect for R-squared results. In addition, LOO computation loo_R() function was updated to use more recent syntax to avoid unnecessary warnings. For the record, the previous version of the case study is available as bayes_R2_v1.
1 Introduction
This notebook demonstrates Bayesian posterior distributions of model based R-squared and LOO-R2 measures for linear and logistic regression. This notebook is a supplement for the paper
- Andrew Gelman, Ben Goodrich, Jonah Gabry, and Aki Vehtari (2018). R-squared for Bayesian regression models. The American Statistician, doi:10.1080/00031305.2018.1549100. Online Preprint.
We specifically define R-squared as \[ R^2 = \frac{{\mathrm Var}_{\mu}}{{\mathrm Var}_{\mu}+{\mathrm Var}_{\mathrm res}}, \] where \({\mathrm Var}_{\mu}\) is variance of modelled predictive means, and \({\mathrm Var}_{\mathrm res}\) is the modelled residual variance. Specifically both of these are computed only using posterior quantities from the fitted model.
The residual based R2 uses draws from the residual distribution and is defined as \[ {\mathrm Var}_{\mu}^s = V_{n=1}^N \hat{y}_n^s\\ {\mathrm Var}_{\mathrm res}^s = V_{n=1}^N \hat{e}_n^s, \] where \(\hat{e}_n^s=y_n-\hat{y}_n^s\).
The model based R2 uses draws from the modeled residual variances. For linear regression we define \[ {\mathrm Var}_{\mathrm res}^s = (\sigma^2)^s, \] and for logistic regression, following Tjur (2009), we define \[ {\mathrm Var}_{\mathrm res}^s = \frac{1}{N}\sum_{n=1}^N (\pi_n^s(1-\pi_n^s)), \] where \(\pi_n^s\) are predicted probabilities.
The LOO-R2 uses LOO residuals and is defined \(1-{\mathrm Var}_{\mathrm loo-res}/{\mathrm Var}_{y}\) and \[ {\mathrm Var}_{y} = V_{n=1}^N {y}_n\\ {\mathrm Var}_{\mathrm loo-res} = V_{n=1}^N \hat{e}_{{\mathrm loo},n}, \] where \(\hat{e}_{{\mathrm loo},n}=y_n-\hat{y}_{{\mathrm loo},n}\). The uncertainty in LOO-R2 comes from not knowing the future data distribution (Vehtari and Ojanen, 2012). We can approximate draws from the corresponding distribution using Bayesian bootstrap (Vehtari and Lampinen, 2002).
File contents:
- bayes_R2_res function (using residuals)
- bayes_R2 function (using sigma and latent space for binary)
- loo_R2 function (using LOO residuals and Bayesian bootstrap for uncertainty)
- code for fitting the models for the example in the paper
- code for producing the plots in the paper (both base graphics and ggplot2 code)
- several examples
Load packages
Data comes from the Regression and Other Stories examples which are included in the rosdata R package, which can be installed with remotes::install_github("avehtari/ROS-Examples",subdir = "rpackage")
library("rosdata")
library("rstanarm")
options(mc.cores = parallel::detectCores())
library("loo")
library("ggplot2")
library("bayesplot")
theme_set(bayesplot::theme_default(base_family = "sans"))
library("latex2exp")
library("foreign")
library("bayesboot")
SEED <- 1800
set.seed(SEED)2 Functions for Bayesian R-squared for stan_glm models.
A more complicated version of this function that is compatible with stan_glmer models and specifying newdata will be available in the rstanarm package.
@param fit A fitted model object returned by stan_glm. @return A vector of R-squared values with length equal to the number of posterior draws.
Bayes-R2 function using residuals
bayes_R2_res <- function(fit) {
y <- rstanarm::get_y(fit)
ypred <- rstanarm::posterior_epred(fit)
if (family(fit)$family == "binomial" && NCOL(y) == 2) {
trials <- rowSums(y)
y <- y[, 1]
ypred <- ypred %*% diag(trials)
}
e <- -1 * sweep(ypred, 2, y)
var_ypred <- apply(ypred, 1, var)
var_e <- apply(e, 1, var)
var_ypred / (var_ypred + var_e)
}Bayes-R2 function using modelled (approximate) residual variance
bayes_R2 <- function(fit) {
mupred <- rstanarm::posterior_epred(fit)
var_mupred <- apply(mupred, 1, var)
if (family(fit)$family == "binomial" && NCOL(y) == 1) {
sigma2 <- apply(mupred*(1-mupred), 1, mean)
} else {
sigma2 <- as.matrix(fit, pars = c("sigma"))^2
}
var_mupred / (var_mupred + sigma2)
}LOO-R2 function using LOO-residuals and Bayesian bootstrap
loo_R2 <- function(fit) {
y <- get_y(fit)
ypred <- posterior_epred(fit)
ll <- log_lik(fit)
M <- length(fit$stanfit@sim$n_save)
N <- fit$stanfit@sim$n_save[[1]]
psis_object <- loo(fit, save_psis=TRUE)$psis_object
ypredloo <- E_loo(ypred, psis_object)$value
eloo <- ypredloo-y
n <- length(y)
rd<-rudirichlet(4000, n)
vary <- (rowSums(sweep(rd, 2, y^2, FUN = "*")) -
rowSums(sweep(rd, 2, y, FUN = "*"))^2)*(n/(n-1))
vareloo <- (rowSums(sweep(rd, 2, eloo^2, FUN = "*")) -
rowSums(sweep(rd, 2, eloo, FUN = "*"))^2)*(n/(n-1))
looR2 <- 1-vareloo/vary
looR2[looR2 < -1] <- -1
looR2[looR2 > 1] <- 1
return(looR2)
}3 Experiments
3.1 Toy data with n=5
x <- 1:5 - 3
y <- c(1.7, 2.6, 2.5, 4.4, 3.8) - 3
xy <- data.frame(x,y)Lsq fit
fit <- lm(y ~ x, data = xy)
ols_coef <- coef(fit)
yhat <- ols_coef[1] + ols_coef[2] * x
r <- y - yhat
rsq_1 <- var(yhat)/(var(y))
rsq_2 <- var(yhat)/(var(yhat) + var(r))
round(c(rsq_1, rsq_2), 3)[1] 0.766 0.766Bayes fit
fit_bayes <- stan_glm(y ~ x, data = xy,
prior_intercept = normal(0, 0.2, autoscale = FALSE),
prior = normal(1, 0.2, autoscale = FALSE),
prior_aux = NULL,
seed = SEED,
refresh = 0
)
posterior <- as.matrix(fit_bayes, pars = c("(Intercept)", "x"))
post_means <- colMeans(posterior)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_bayes)), 2)[1] 0.8round(median(bayesR2<-bayes_R2(fit_bayes)), 2)[1] 0.74LOO-R2
looR2<-loo_R2(fit_bayes)
round(median(looR2), 2)[1] 0.44With small n LOO-R2 is much lower than median of Bayesian R2
Figures
The first section of code below creates plots using base R graphics. Below that there is code to produce the plots using ggplot2.
# take a sample of 20 posterior draws
keep <- sample(nrow(posterior), 20)
samp_20_draws <- posterior[keep, ]Base graphics version
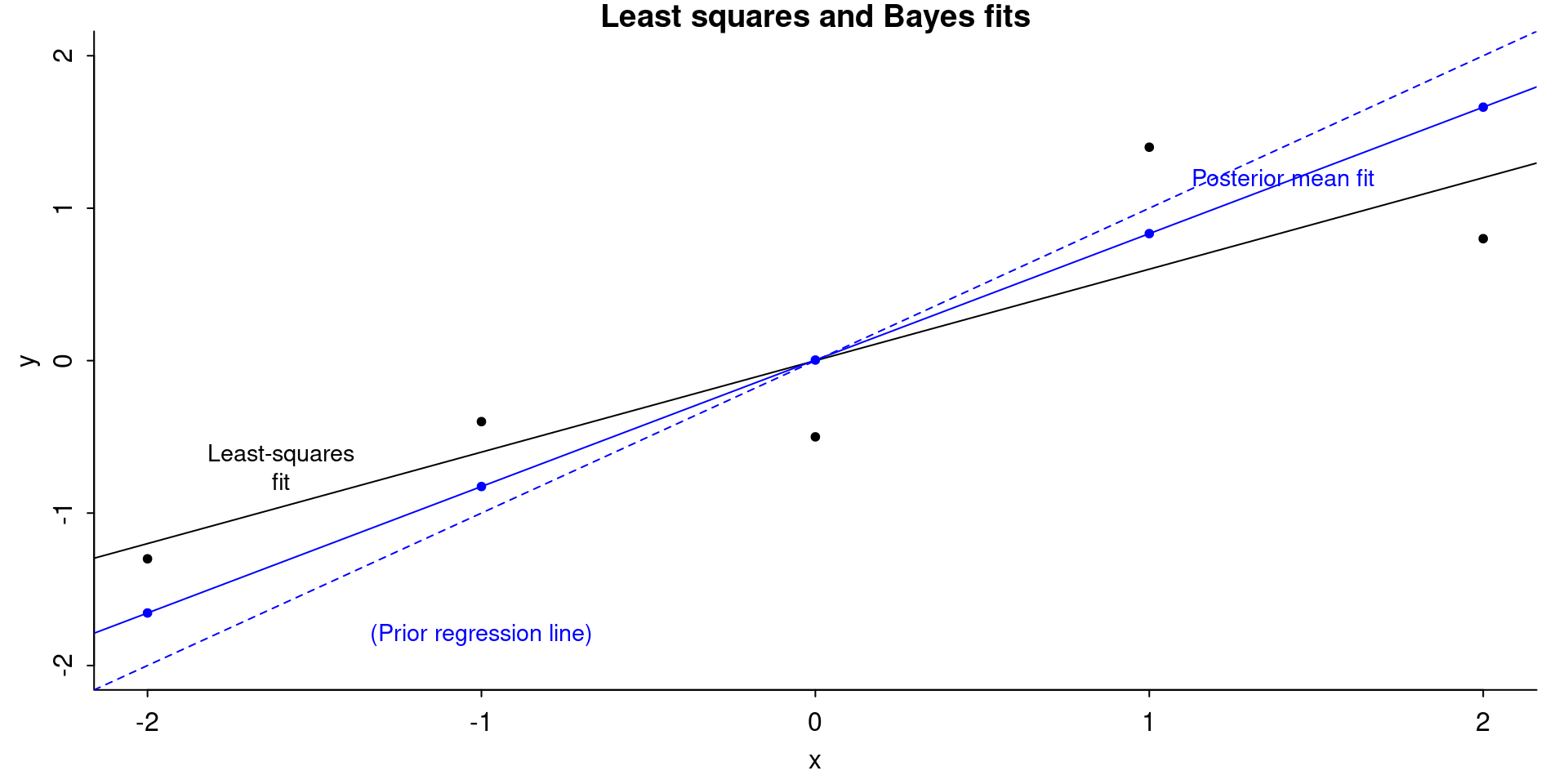
par(mar=c(3,3,1,1), mgp=c(1.7,.5,0), tck=-.01)
plot(
x, y,
ylim = range(x),
xlab = "x",
ylab = "y",
main = "Least squares and Bayes fits",
bty = "l",
pch = 20
)
abline(coef(fit)[1], coef(fit)[2], col = "black")
text(-1.6,-.7, "Least-squares\nfit", cex = .9)
abline(0, 1, col = "blue", lty = 2)
text(-1, -1.8, "(Prior regression line)", col = "blue", cex = .9)
abline(coef(fit_bayes)[1], coef(fit_bayes)[2], col = "blue")
text(1.4, 1.2, "Posterior mean fit", col = "blue", cex = .9)
points(
x,
coef(fit_bayes)[1] + coef(fit_bayes)[2] * x,
pch = 20,
col = "blue"
)
par(mar=c(3,3,1,1), mgp=c(1.7,.5,0), tck=-.01)
plot(
x, y,
ylim = range(x),
xlab = "x",
ylab = "y",
bty = "l",
pch = 20,
main = "Bayes posterior simulations"
)
for (s in 1:nrow(samp_20_draws)) {
abline(samp_20_draws[s, 1], samp_20_draws[s, 2], col = "#9497eb")
}
abline(
coef(fit_bayes)[1],
coef(fit_bayes)[2],
col = "#1c35c4",
lwd = 2
)
points(x, y, pch = 20, col = "black")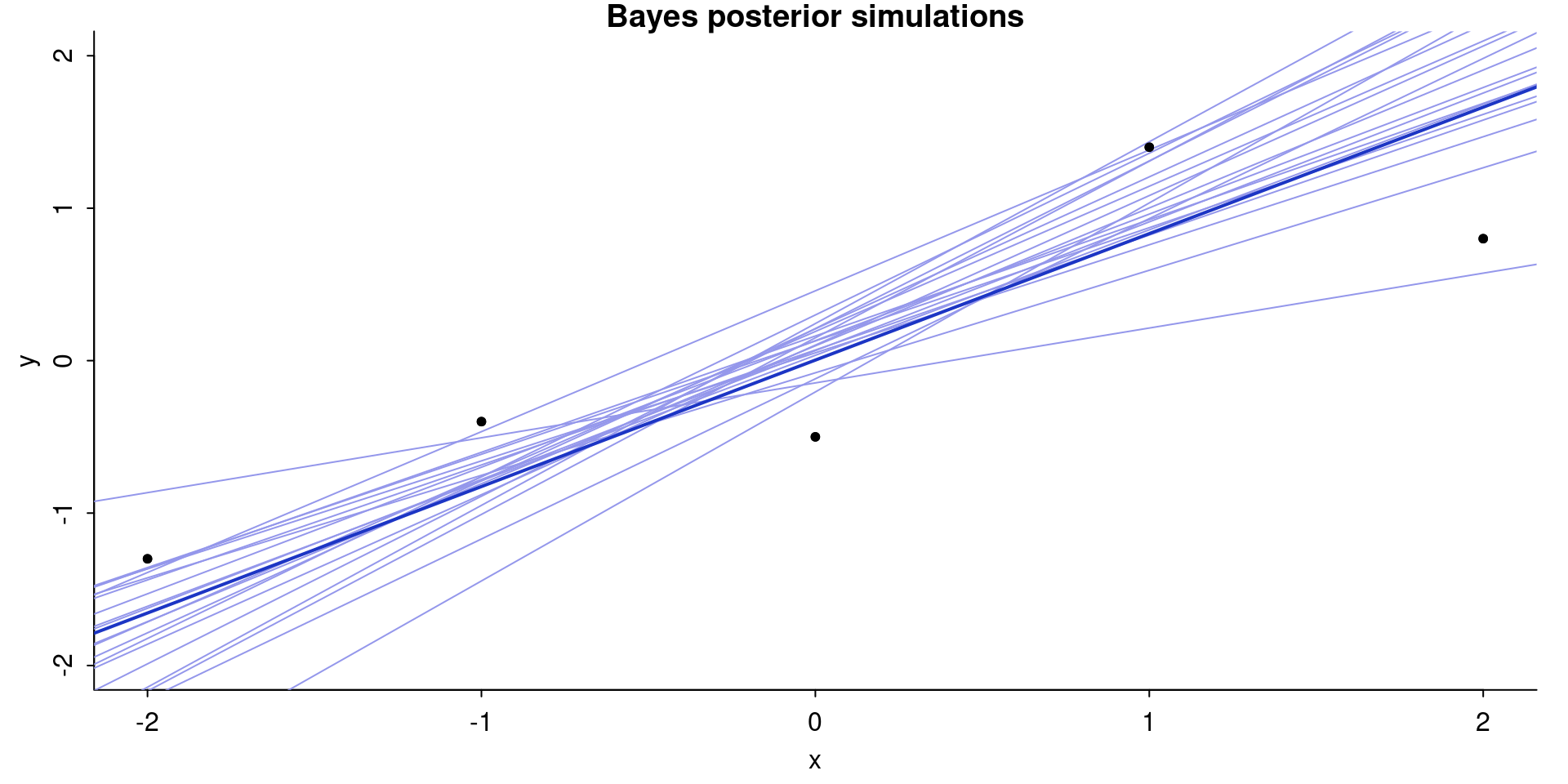
ggplot version
theme_update(
plot.title = element_text(face = "bold", hjust = 0.5),
axis.text = element_text(size = rel(1.1))
)
fig_1a <-
ggplot(xy, aes(x, y)) +
geom_point() +
geom_abline( # ols regression line
intercept = ols_coef[1],
slope = ols_coef[2],
size = 0.4
) +
geom_abline( # prior regression line
intercept = 0,
slope = 1,
color = "blue",
linetype = 2,
size = 0.3
) +
geom_abline( # posterior mean regression line
intercept = post_means[1],
slope = post_means[2],
color = "blue",
size = 0.4
) +
geom_point(
aes(y = post_means[1] + post_means[2] * x),
color = "blue"
) +
annotate(
geom = "text",
x = c(-1.6, -1, 1.4),
y = c(-0.7, -1.8, 1.2),
label = c(
"Least-squares\nfit",
"(Prior regression line)",
"Posterior mean fit"
),
color = c("black", "blue", "blue"),
size = 3.8
) +
ylim(range(x)) +
ggtitle("Least squares and Bayes fits")
plot(fig_1a)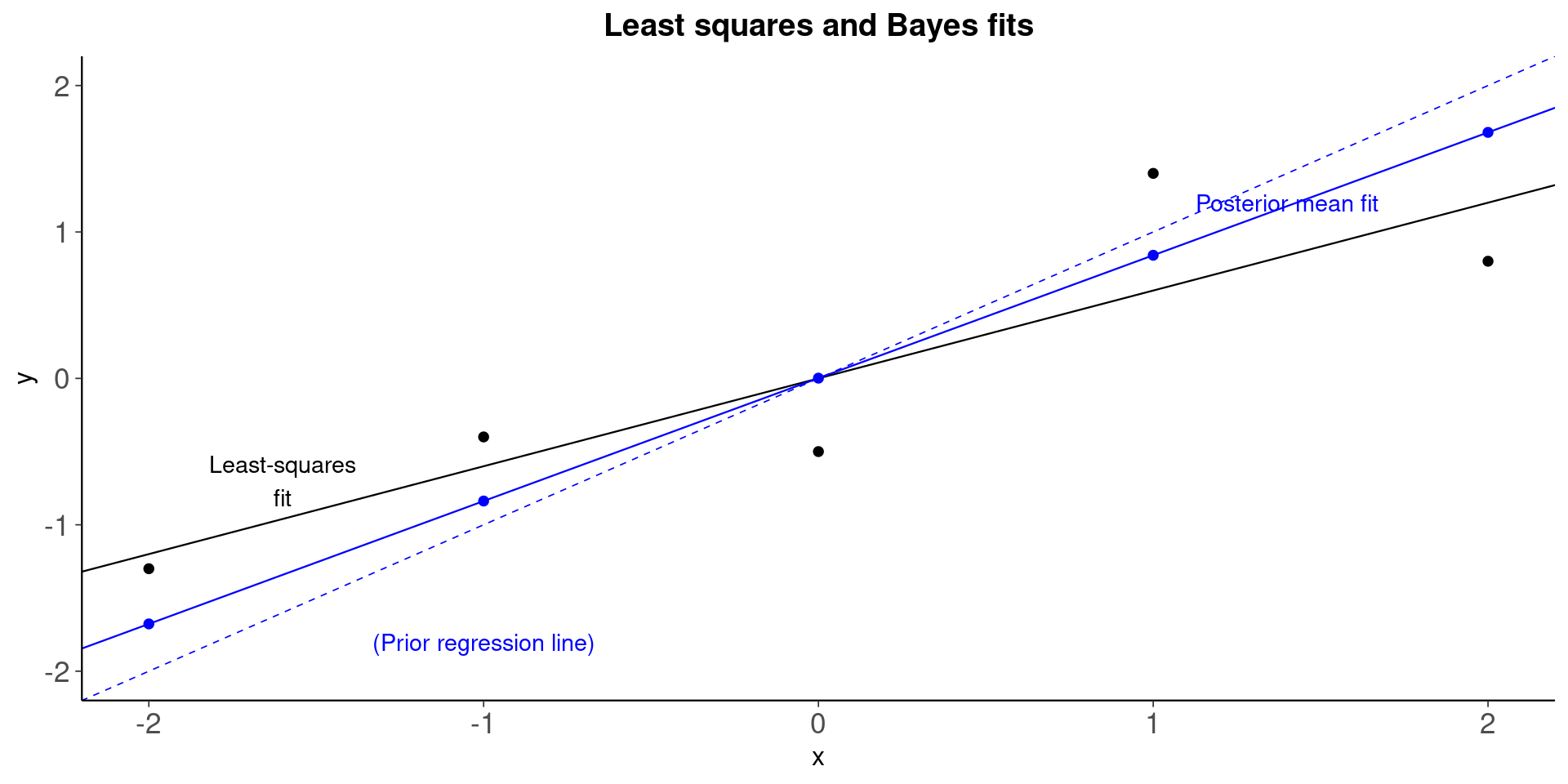
fig_1b <-
ggplot(xy, aes(x, y)) +
geom_abline( # 20 posterior draws of the regression line
intercept = samp_20_draws[, 1],
slope = samp_20_draws[, 2],
color = "#9497eb",
size = 0.25
) +
geom_abline( # posterior mean regression line
intercept = post_means[1],
slope = post_means[2],
color = "#1c35c4",
size = 1
) +
geom_point() +
ylim(range(x)) +
ggtitle("Bayes posterior simulations")
plot(fig_1b)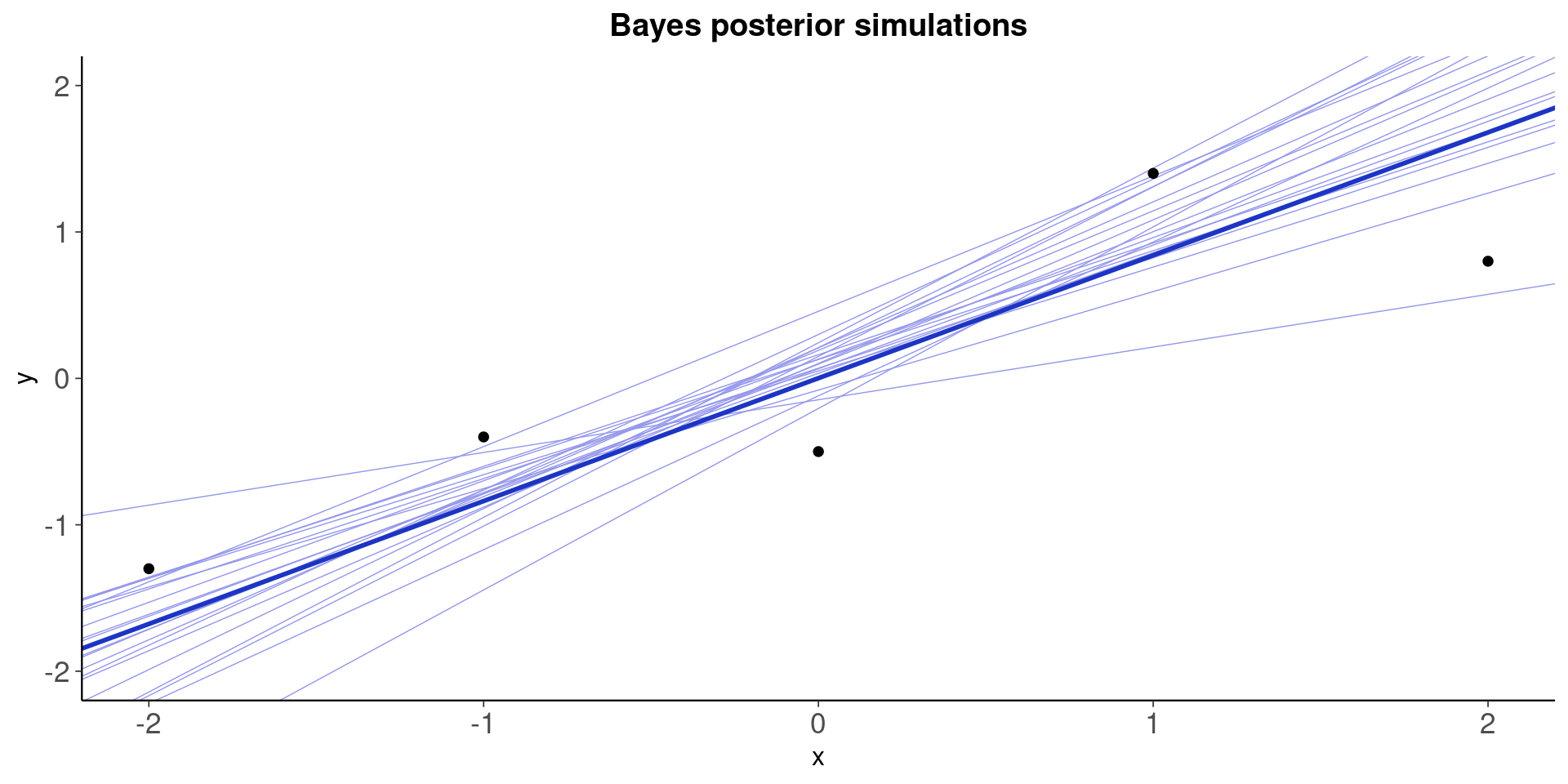
Bayesian R^2 Posterior and median
mcmc_hist(data.frame(bayesR2), binwidth=0.02)+xlim(c(0,1))+
xlab(TeX('$R^2$'))+
geom_vline(xintercept=median(bayesR2))+
ggtitle('Bayesian R squared posterior and median')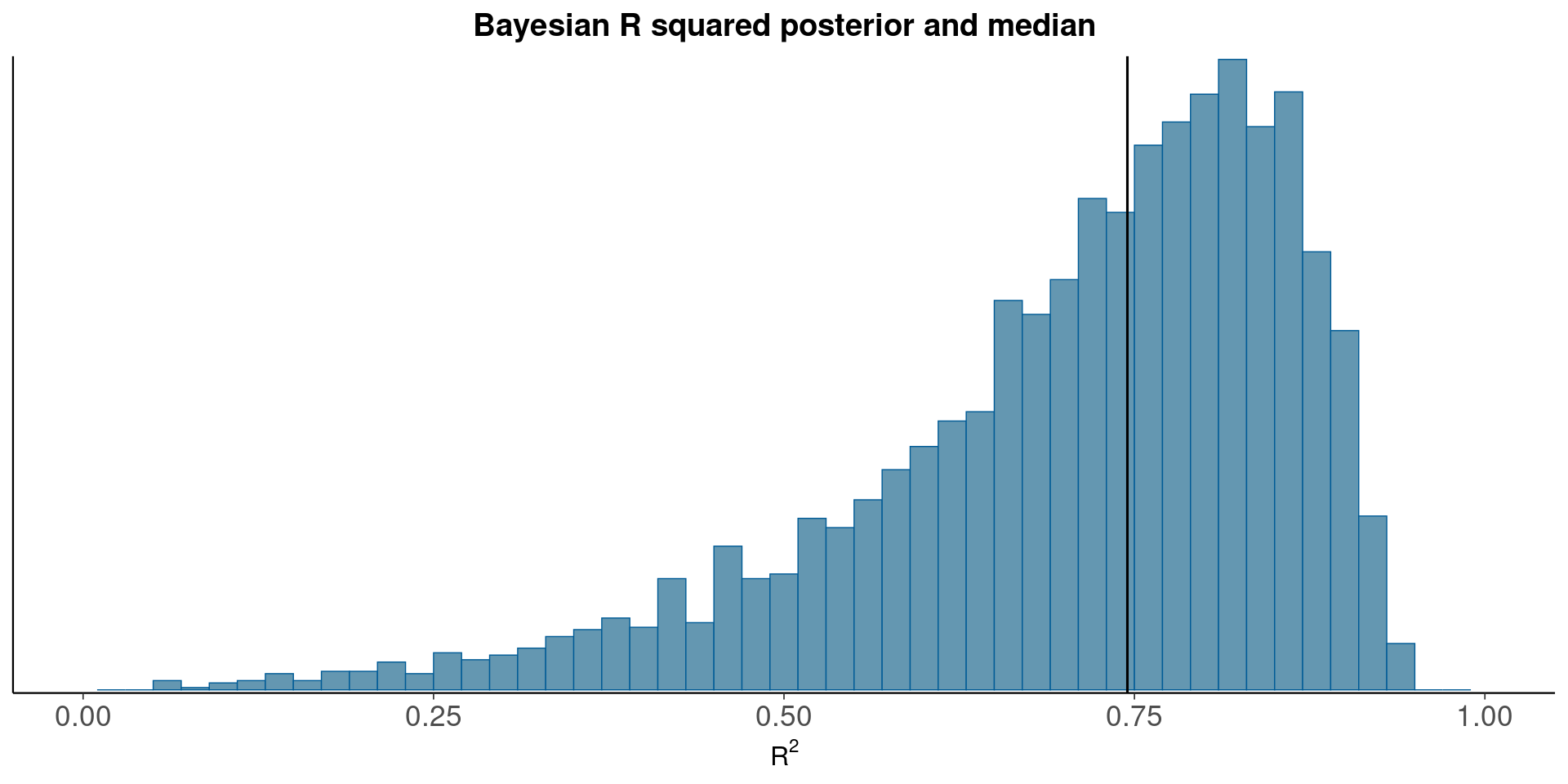
Compare draws from residual variances and sigma
fit<-fit_bayes
y <- rstanarm::get_y(fit)
ypred <- rstanarm::posterior_epred(fit)
e <- -1 * sweep(ypred, 2, y)
var_e <- apply(e, 1, var)
sigma <- as.matrix(fit, pars = c("sigma"))
qplot(sqrt(var_e),sigma)+geom_abline()+
ggtitle('Toy data with n=5')+
xlab('Draws from residual sd')+
ylab('Draws from sigma')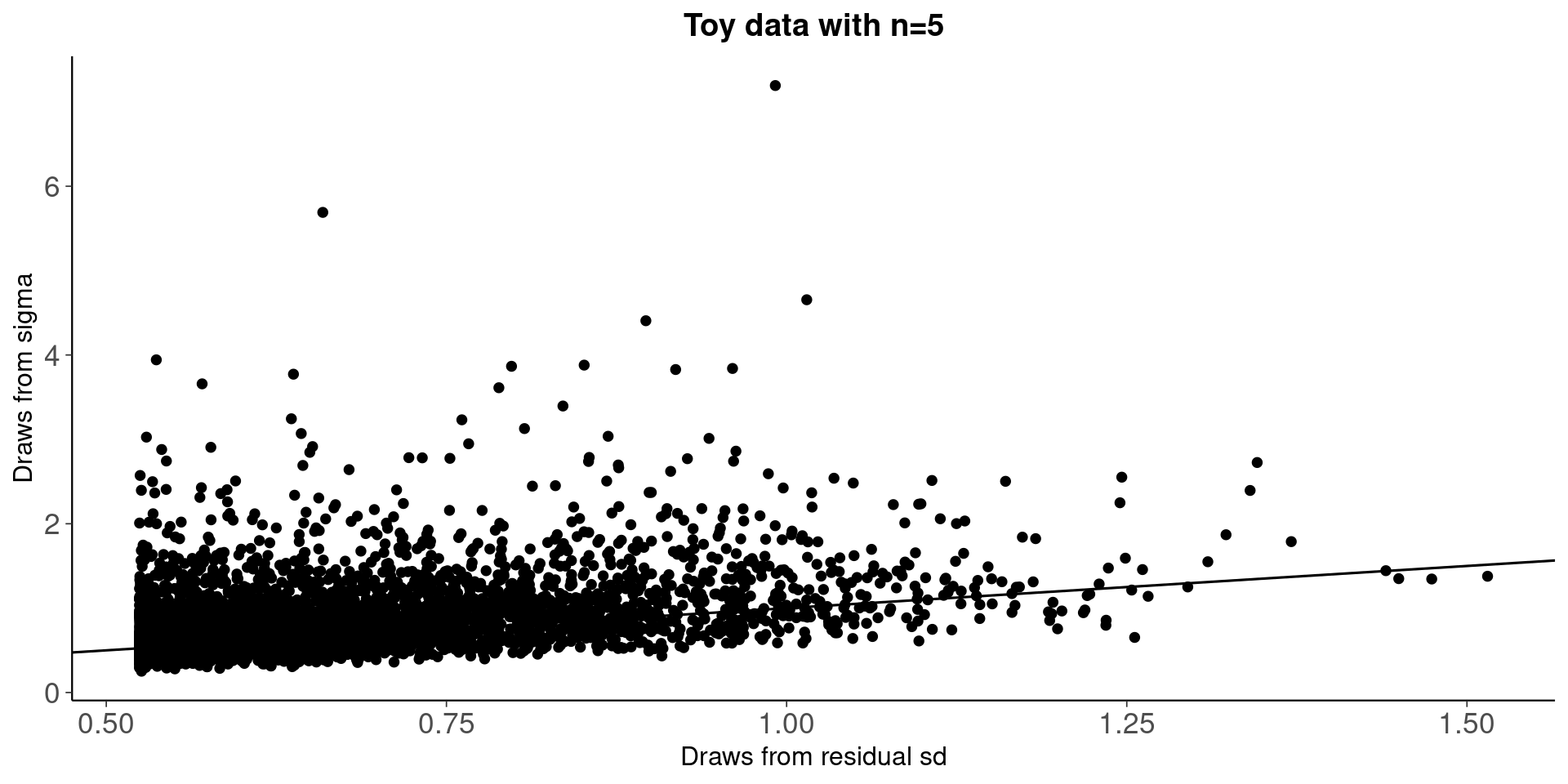
Draws from residual sd are truncated and not properly using the model information, which is reflected in R2 distribution below.
qplot(bayes_R2_res(fit_bayes),bayes_R2(fit_bayes))+geom_abline()+
ggtitle('Toy data with n=5')+
xlab('Bayesian R2 with draws from residual variance')+
ylab('Bayesian R2 with draws from sigma')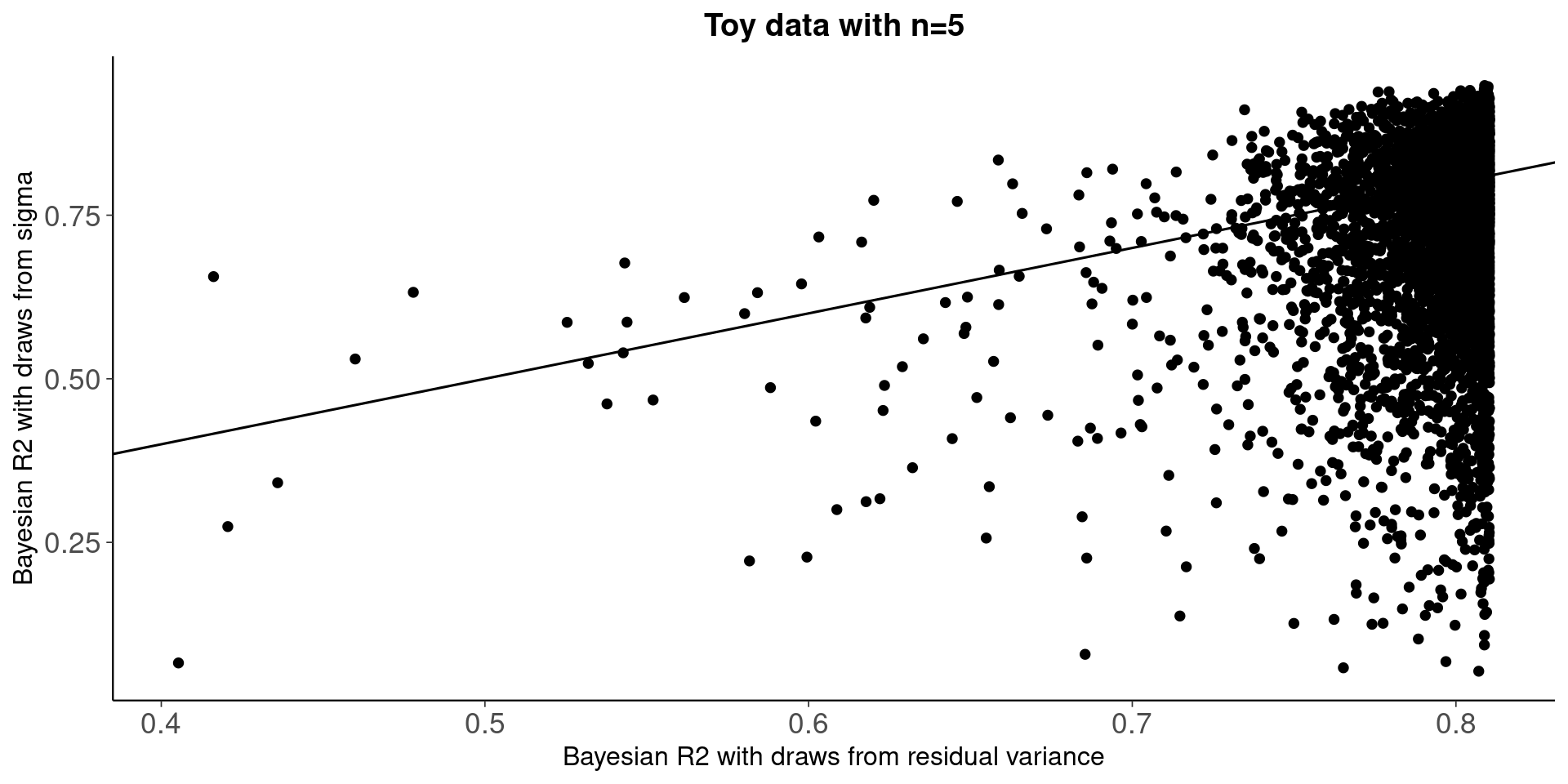
Compare residual and model based Bayesian R2, and Bayesian LOO-R2
pxl<-xlim(-0.5,1)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.02)+pxl+
ggtitle('Toy data with n=5')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.02)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.02)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)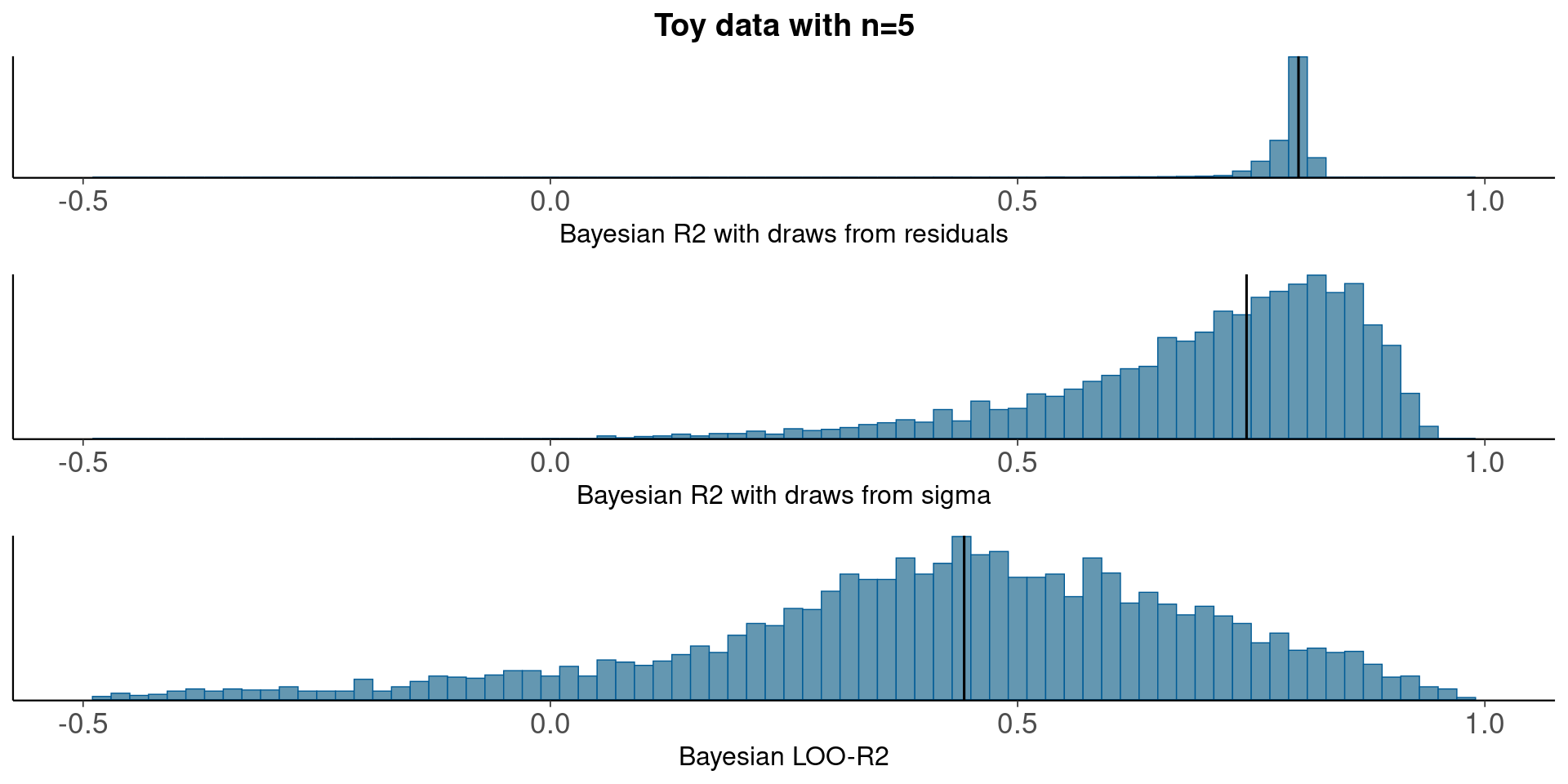
With small n, draws from the residual variance are not a good representation of the uncertainty.
3.2 Toy logistic regression example, n=20
set.seed(20)
y<-rbinom(n=20,size=1,prob=(1:20-0.5)/20)
data <- data.frame(rvote=y, income=1:20)fit_logit <- stan_glm(rvote ~ income, family=binomial(link="logit"), data=data,
refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_logit)), 2)[1] 0.36round(median(bayesR2<-bayes_R2(fit_logit)), 2)[1] 0.38LOO-R2
looR2<-loo_R2(fit_logit)
round(median(looR2), 2)[1] 0.26With small n LOO-R2 is much lower than median of Bayesian R2
Compare residual and model based Bayesian R2, and Bayesian LOO-R2
pxl<-xlim(-.5,1)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.05)+pxl+
ggtitle('Toy logistic data with n=20')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.05)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.05)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)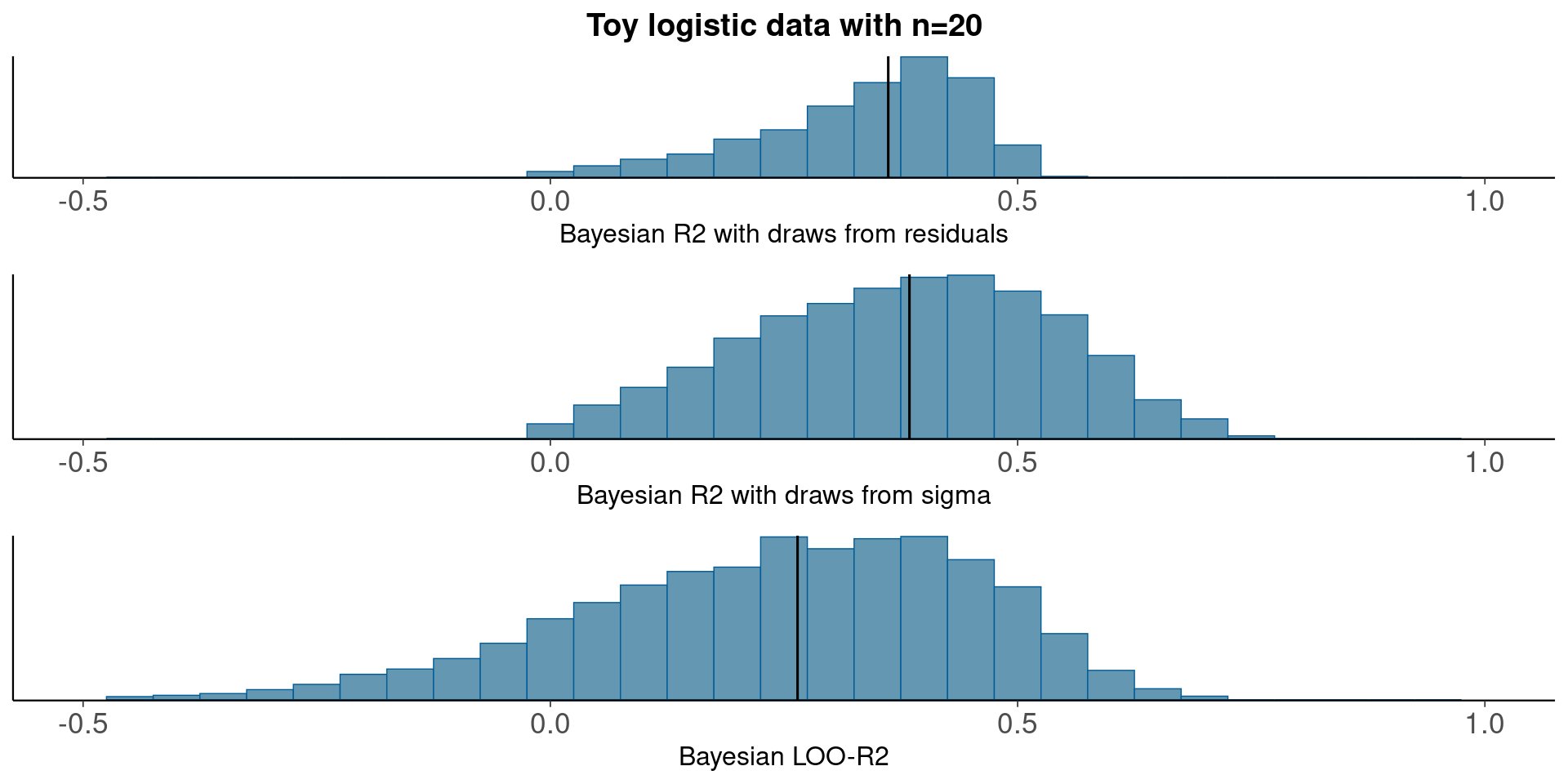
With small n, draws from the residual variance are not a good representation of the uncertainty.
3.3 Mesquite - linear regression
Predicting the yields of mesquite bushes, n=46
Prepare data
head(mesquite) obs group diam1 diam2 total_height canopy_height density weight canopy_volume canopy_area
1 1 MCD 1.8 1.15 1.30 1.00 1 401.3 2.07000 2.070
2 2 MCD 1.7 1.35 1.35 1.33 1 513.7 3.05235 2.295
3 3 MCD 2.8 2.55 2.16 0.60 1 1179.2 4.28400 7.140
4 4 MCD 1.3 0.85 1.80 1.20 1 308.0 1.32600 1.105
5 5 MCD 3.3 1.90 1.55 1.05 1 855.2 6.58350 6.270
6 6 MCD 1.4 1.40 1.20 1.00 1 268.7 1.96000 1.960
canopy_shape
1 1.565217
2 1.259259
3 1.098039
4 1.529412
5 1.736842
6 1.000000mesquite$canopy_volume <- mesquite$diam1 * mesquite$diam2 * mesquite$canopy_height
mesquite$canopy_area <- mesquite$diam1 * mesquite$diam2
mesquite$canopy_shape <- mesquite$diam1 / mesquite$diam2
(n <- nrow(mesquite))[1] 46Predict log weight model with log canopy volume, log canopy shape, and group
fit_5 <- stan_glm(log(weight) ~ log(canopy_volume) + log(canopy_shape) + group,
data=mesquite, refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_5)), 2)[1] 0.87round(median(bayesR2<-bayes_R2(fit_5)), 2)[1] 0.87LOO-R2
looR2<-loo_R2(fit_5)
round(median(looR2), 2)[1] 0.85LOO-R2 is slightly lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(0.6, 0.95)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.01)+pxl+
ggtitle('Mesquite data with n=46')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.01)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.01)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)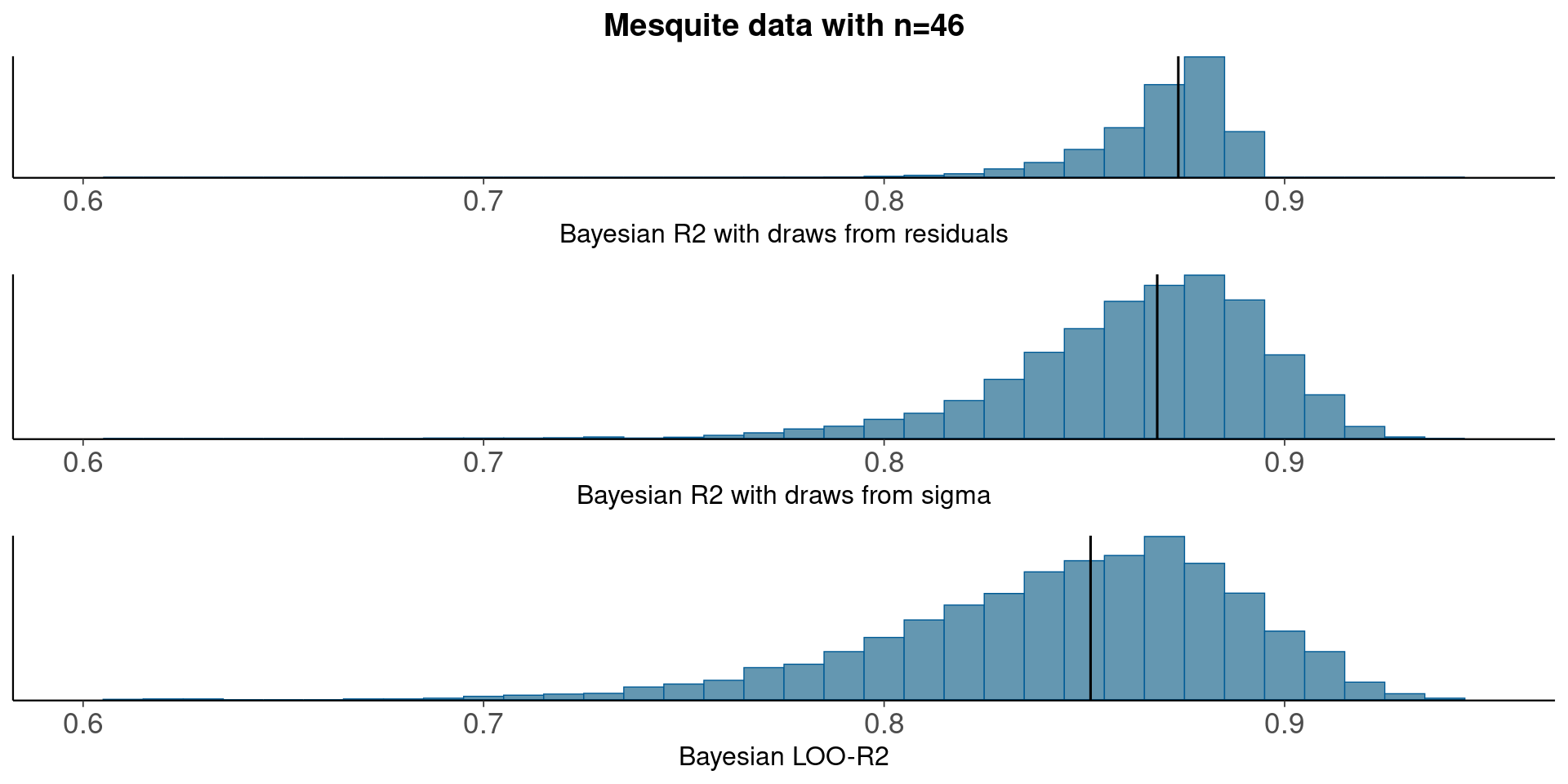
With small n, draws from the residual variance are not a good representation of the uncertainty and R2 distribution is more narrow (50%) than when using draws from the model sigma.
3.4 LowBwt – logistic regression
Predict low birth weight, n=189, from Hosmer and Lemeshow (2000) This data was also used by Tjur (2009).
Prepare data
head(lowbwt) ID low age lwt race smoke ptl ht ui ftv bwt
1 85 0 19 182 2 0 0 0 1 0 2523
2 86 0 33 155 3 0 0 0 0 3 2551
3 87 0 20 105 1 1 0 0 0 1 2557
4 88 0 21 108 1 1 0 0 1 2 2594
5 89 0 18 107 1 1 0 0 1 0 2600
6 91 0 21 124 3 0 0 0 0 0 2622lowbwt$race <- factor(lowbwt$race)
(n <- nrow(lowbwt))[1] 189Predict low birth weight
fit <- stan_glm(low ~ age + lwt + race + smoke,
family=binomial(link="logit"), data=lowbwt, refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit)), 2)[1] 0.12round(median(bayesR2<-bayes_R2(fit)), 2)[1] 0.12LOO-R2
looR2<-loo_R2(fit)
round(median(looR2), 2)[1] 0.03LOO-R2 is much lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(-0.15, 0.3)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.01)+pxl+
ggtitle('Low birth weight data with n=189')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.01)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.01)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)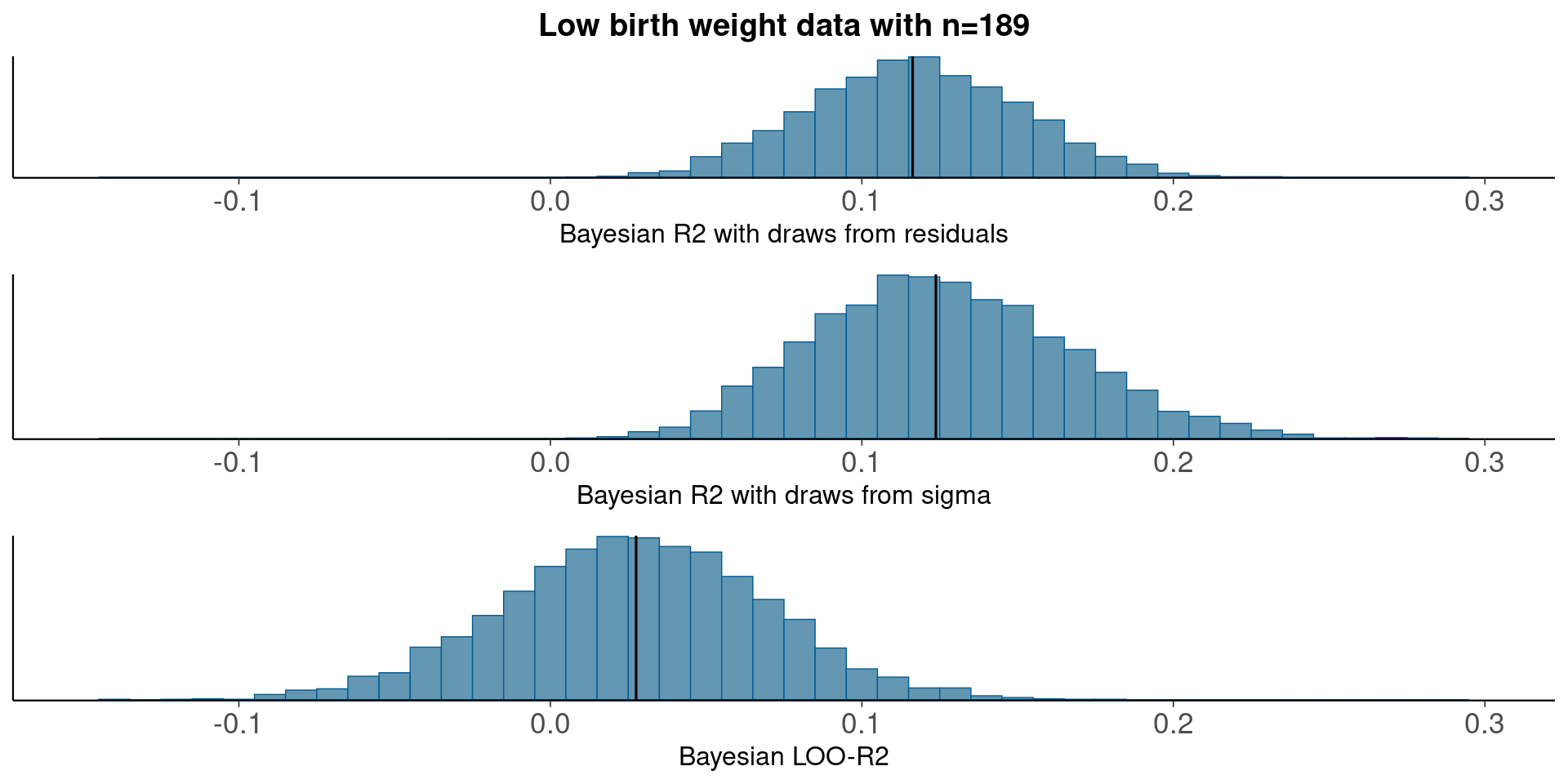
With moderate n, the distribution obtained using draws from the residual variances is still more narrow than the distribution obtained using draws from the model based variance.
3.5 LowBwt – linear regression
Predict birth weight, n=189, from Hosmer and Lemeshow (2000). Tjur (2009) used logistic regression for dichotomized birth weight. Below we use the continuos valued birth weight.
Predict birth weight
fit <- stan_glm(bwt ~ age + lwt + race + smoke, data=lowbwt, refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit)), 2)[1] 0.16round(median(bayesR2<-bayes_R2(fit)), 2)[1] 0.16LOO-R2
looR2<-loo_R2(fit)
round(median(looR2), 2)[1] 0.09LOO-R2 is much lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(-0.12, 0.36)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.01)+pxl+
ggtitle('Birth weight data with n=189')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.01)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.01)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)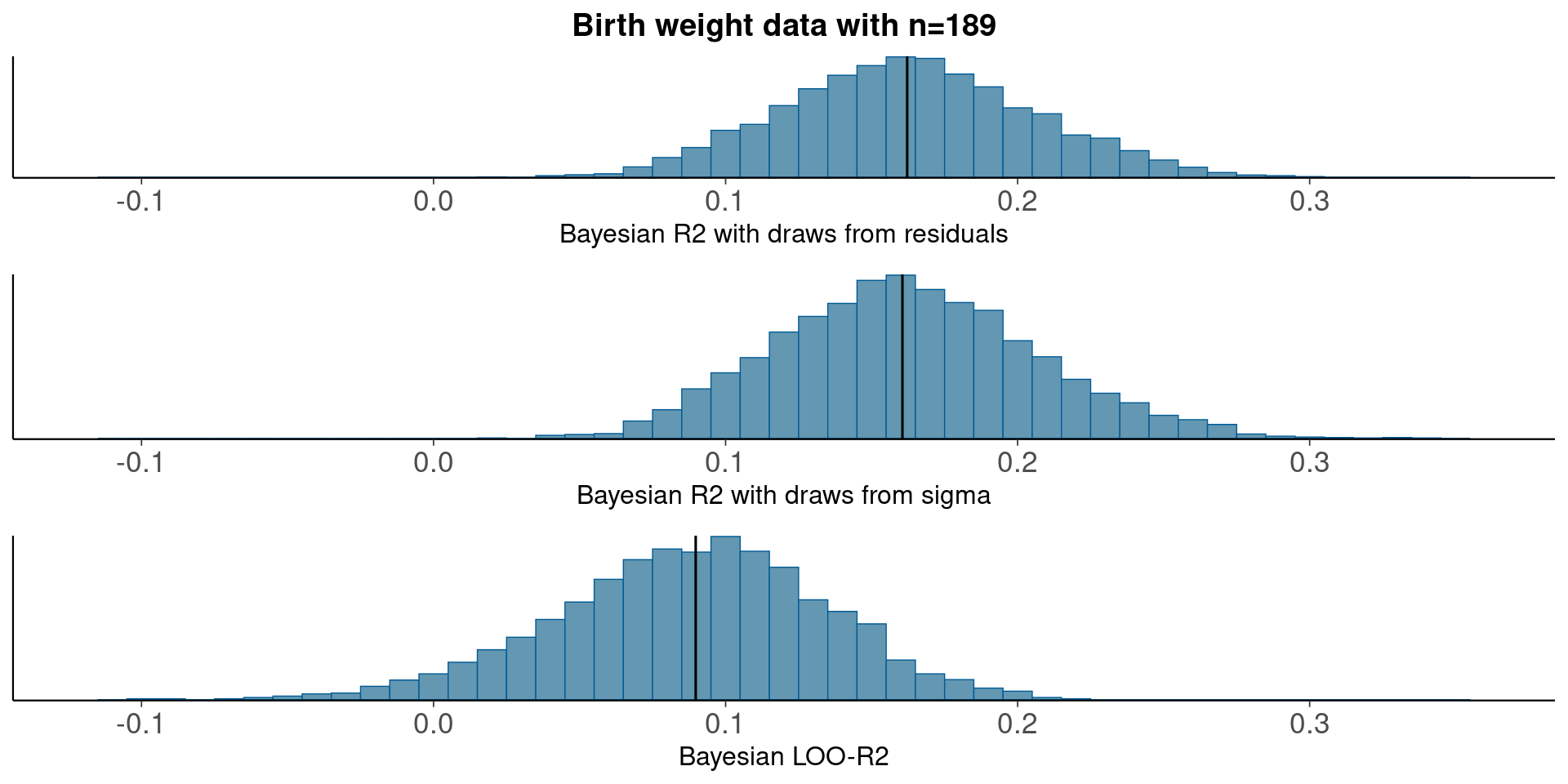
With moderate n, the distribution obtained using draws from the residual variances is still more narrow than the distribution obtained using draws from the model based variance.
3.6 KidIQ - linear regression
Children’s test scores data, n=434
Prepare data
head(kidiq) kid_score mom_hs mom_iq mom_age
1 65 1 121.11753 27
2 98 1 89.36188 25
3 85 1 115.44316 27
4 83 1 99.44964 25
5 115 1 92.74571 27
6 98 0 107.90184 18(n <- nrow(kidiq))[1] 434Predict test score
fit_3 <- stan_glm(kid_score ~ mom_hs + mom_iq, data=kidiq,
seed=1507, refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_3)), 2)[1] 0.22round(median(bayesR2<-bayes_R2(fit_3)), 2)[1] 0.21LOO-R2
looR2<-loo_R2(fit_3)
round(median(looR2), 2)[1] 0.2LOO-R2 is slightly lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(0.05, 0.35)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.01)+pxl+
ggtitle('KidIQ data with n=434')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.01)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.01)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)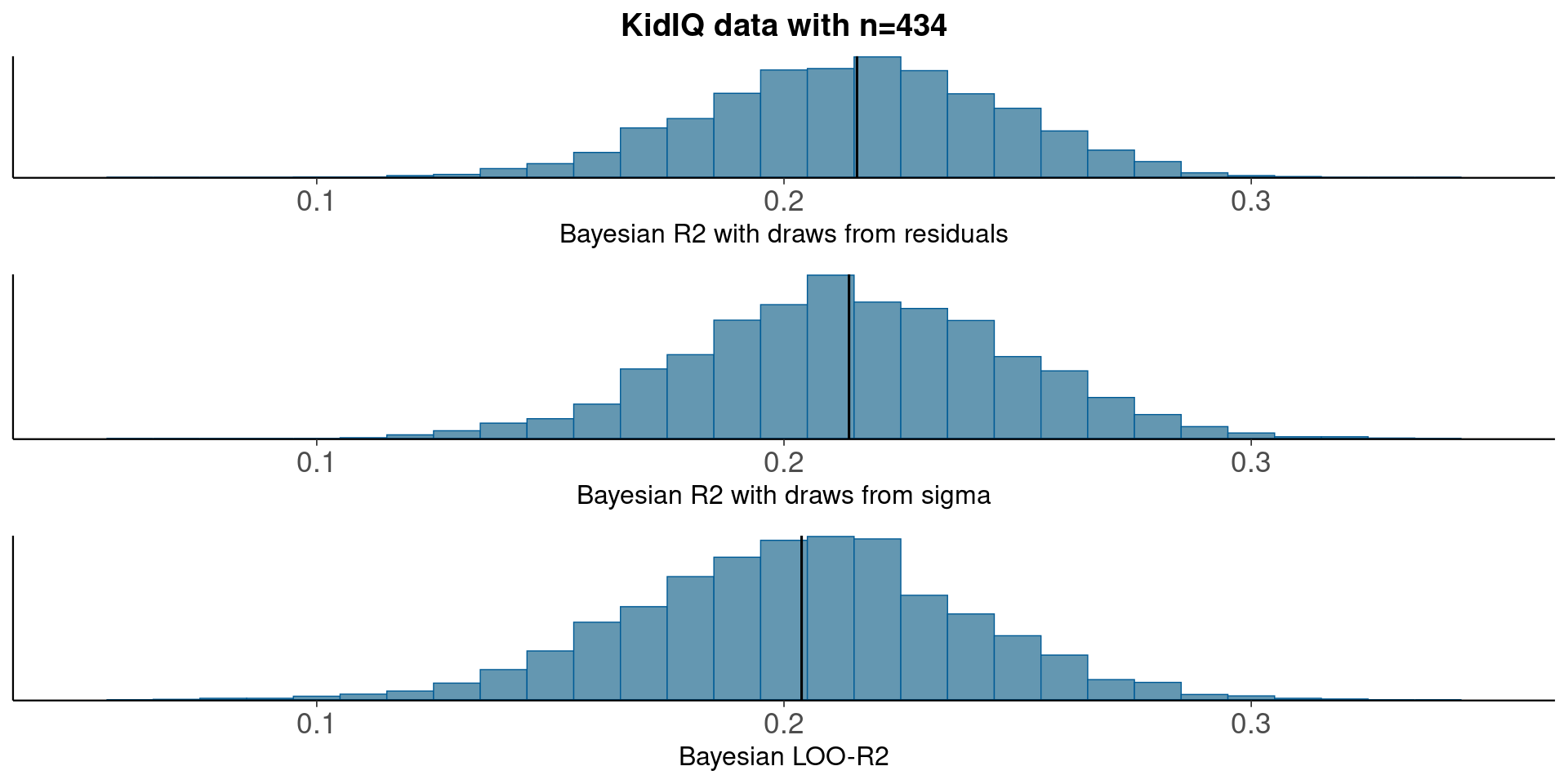
3.7 Earnings - logistic and linear regression
Predict respondents’ yearly earnings using survey data from 1990. logistic regression n=1374, linear regression n=1187
Prepare data
head(earnings) height weight male earn earnk ethnicity education mother_education father_education walk
1 74 210 1 50000 50 White 16 16 16 3
2 66 125 0 60000 60 White 16 16 16 6
3 64 126 0 30000 30 White 16 16 16 8
4 65 200 0 25000 25 White 17 17 NA 8
5 63 110 0 50000 50 Other 16 16 16 5
6 68 165 0 62000 62 Black 18 18 18 1
exercise smokenow tense angry age positive log_earn
1 3 2 0 0 45 TRUE 10.81978
2 5 1 0 0 58 TRUE 11.00210
3 1 2 1 1 29 TRUE 10.30895
4 1 2 0 0 57 TRUE 10.12663
5 6 2 0 0 91 TRUE 10.81978
6 1 2 2 2 54 TRUE 11.03489earnings_all <- earnings
earnings_all$positive <- earnings_all$earn > 0
(n_all <- nrow(earnings_all))[1] 1629# only non-zero earnings
earnings <- earnings_all[earnings_all$positive, ]
(n <- nrow(earnings))[1] 1629earnings$log_earn <- log(earnings$earn)
earnings_all$positive <- as.numeric(earnings_all$positive)Bayesian logistic regression on non-zero earnings Null model
fit_0 <- stan_glm(positive ~ 1, family = binomial(link = "logit"),
data = earnings_all, refresh=0)loo0<-loo(fit_0)Predict using height and sex
fit_1a <- stan_glm(positive ~ height + male, family = binomial(link = "logit"),
data = earnings_all, refresh=0)loo1a<-loo(fit_1a)
loo_compare(loo0, loo1a) elpd_diff se_diff
fit_0 0.0 0.0
fit_1a 0.0 0.0 There is a clear difference in predictive performance.
Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_1a)), 3)[1] 0.5round(median(bayesR2<-bayes_R2(fit_1a)), 3)[1] 0.001LOO-R2
looR2<-loo_R2(fit_1a)
round(median(looR2), 2)[1] -1LOO-R2 is slightly lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(0.02, 0.11)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.002)+pxl+
ggtitle('Earnings data with n=1374')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.002)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.002)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)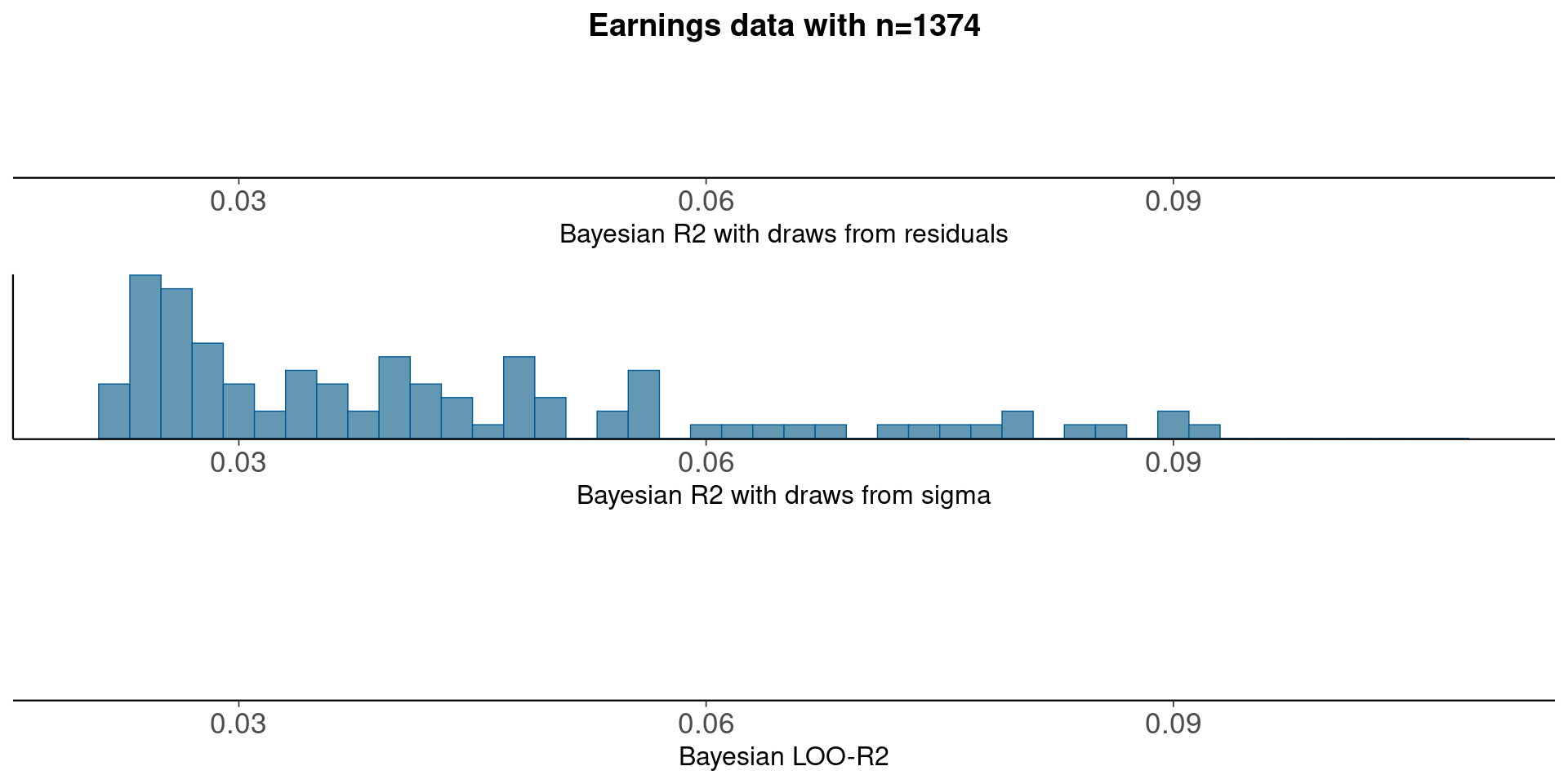
With plenty of data, there is not much difference between using draws from residuals or from sigma.
Bayesian probit regression on non-zero earnings
fit_1p <- stan_glm(positive ~ height + male,
family = binomial(link = "probit"),
data = earnings_all, refresh=0)loo1p<-loo(fit_1p)Warning: Found 7 observation(s) with a pareto_k > 0.7. We recommend calling 'loo' again with argument 'k_threshold = 0.7' in order to calculate the ELPD without the assumption that these observations are negligible. This will refit the model 7 times to compute the ELPDs for the problematic observations directly.loo_compare(loo1a, loo1p) elpd_diff se_diff
fit_1p 0.0 0.0
fit_1a -1.0 0.0 There is no practical difference in predictive performance between logit and probit.
Median Bayesian R2
round(median(bayesR2), 3)[1] 0.001round(median(bayesR2p<-bayes_R2(fit_1p)), 3)[1] 0LOO-R2
looR2p<-loo_R2(fit_1p)Warning: Found 7 observation(s) with a pareto_k > 0.7. We recommend calling 'loo' again with argument 'k_threshold = 0.7' in order to calculate the ELPD without the assumption that these observations are negligible. This will refit the model 7 times to compute the ELPDs for the problematic observations directly.round(median(looR2), 2)[1] -1round(median(looR2p), 2)[1] -1LOO-R2 is slightly lower than median of Bayesian R2
Compare logistic and probit models using new Bayesian R2
pxl<-xlim(0.02, 0.11)
p1<-mcmc_hist(data.frame(bayesR2), binwidth=0.002)+pxl+
ggtitle('Earnings data with n=1374')+
xlab('Bayesian R2 for logistic model')+
geom_vline(xintercept=median(bayesR2))
p2<-mcmc_hist(data.frame(bayesR2p), binwidth=0.002)+pxl+
xlab('Bayesian R2 for probit model')+
geom_vline(xintercept=median(bayesR2p))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.002)+pxl+
xlab('Bayesian LOO-R2 for logistic model')+
geom_vline(xintercept=median(looR2))
p4<-mcmc_hist(data.frame(looR2p), binwidth=0.002)+pxl+
xlab('Bayesian LOO-R2 for probit model')+
geom_vline(xintercept=median(looR2p))
bayesplot_grid(p1,p3,p2,p4)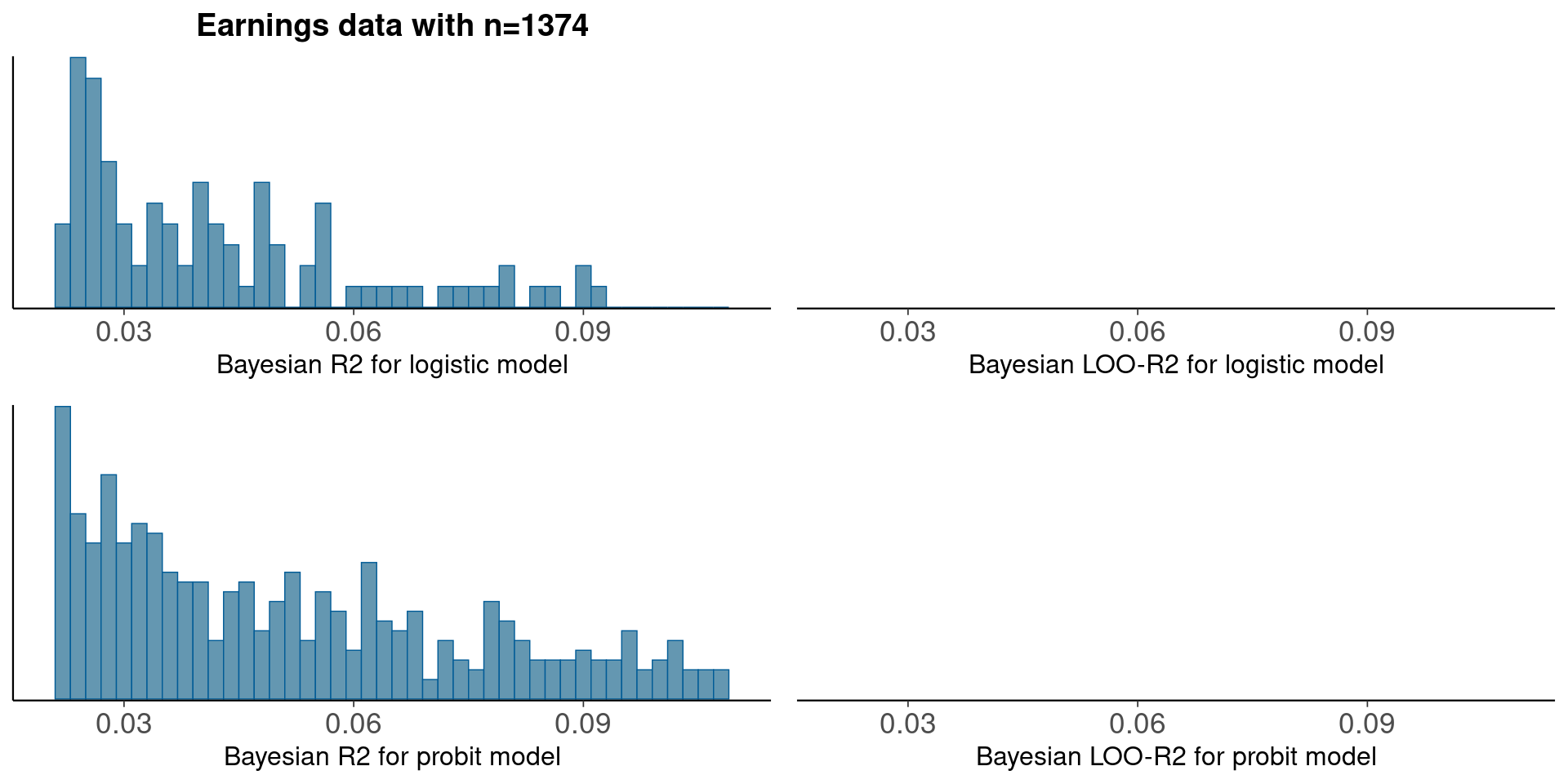
There is no practical difference in predictive performance between logit and probit.
Bayesian model on positive earnings on log scale
fit_1b <- stan_glm(log_earn ~ height + male, data = earnings, refresh=0)Median Bayesian R2
round(median(bayesR2res<-bayes_R2_res(fit_1b)), 3)[1] 0.08round(median(bayesR2<-bayes_R2(fit_1b)), 3)[1] 0.08LOO-R2
looR2<-loo_R2(fit_1b)
round(median(looR2), 2)[1] 0.08LOO-R2 is slightly lower than median of Bayesian R2
Compare residual and model based Bayesian R2
pxl<-xlim(0.02, 0.15)
p1<-mcmc_hist(data.frame(bayesR2res), binwidth=0.002)+pxl+
ggtitle('Positive earnings data with n=1187')+
xlab('Bayesian R2 with draws from residuals')+
geom_vline(xintercept=median(bayesR2res))
p2<-mcmc_hist(data.frame(bayesR2), binwidth=0.002)+pxl+
xlab('Bayesian R2 with draws from sigma')+
geom_vline(xintercept=median(bayesR2))
p3<-mcmc_hist(data.frame(looR2), binwidth=0.002)+pxl+
xlab('Bayesian LOO-R2')+
geom_vline(xintercept=median(looR2))
bayesplot_grid(p1,p2,p3)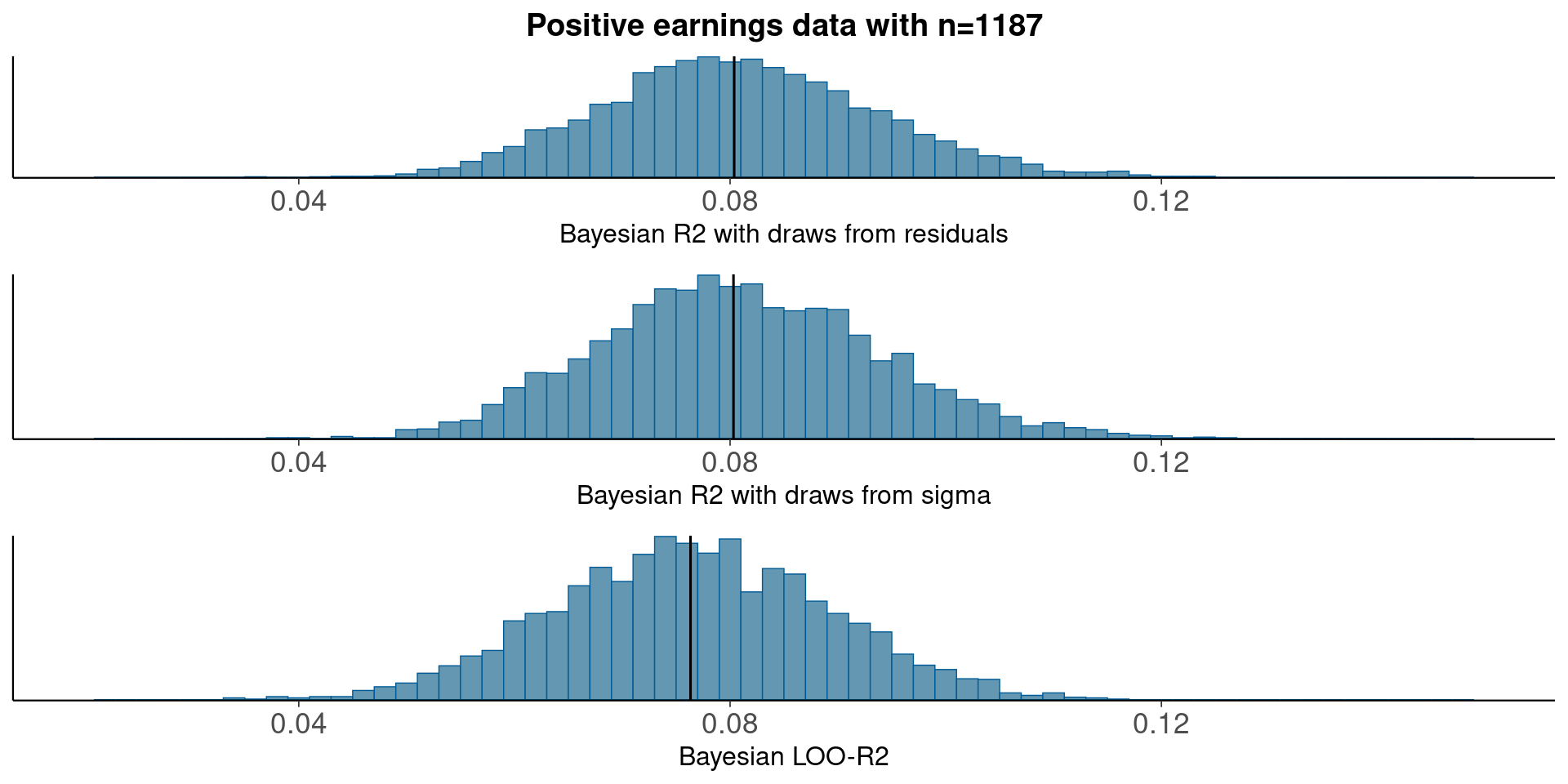
With plenty of data, there is not much difference between using draws from residuals or from sigma.
References
Hosmer, D. W. and Lemeshow, S. (2000) Applied logistic regression. Wiley.
Tjur, T. (2009) ‘Coefficient of determination in logistic regression models—A new proposal: The coefficient of discrimination’, American Statistician, 63, pp. 366–372.
Vehtari, A. and Lampinen, J. (2002) ‘Bayesian model assessment and comparison using cross-validation predictive densities’, Neural Computation, 14(10), pp. 2439–2468.
Vehtari, A. and Ojanen, J. (2012) ‘A survey of Bayesian predictive methods for model assessment, selection and comparison’, Statistics Surveys, 6, pp. 142–228.